Abstract
Introduction
The microenvironment of cells around multiple myeloma (MM) tumors strongly influences patient outcomes, as evidenced by the success of immunomodulatory therapies. To develop precision immunotherapeutic approaches, it is essential to identify and enumerate pro- and anti-myelomatous cell types within the bone marrow.
Methods
The population of immune and other non-tumor cell types was estimated from RNA microarray data derived from paired whole bone marrow (WBM) core biopsies and CD138-enriched (CD138+) bone marrow aspirates at presentation from 405 newly diagnosed MM patients who underwent similar treatments. Expression profiles from CD138+ cells were used to infer the residual transcriptome contribution of immune and stromal cells in the WBM array data. We performed unsupervised clustering of the estimated microenvironment gene expression profiles and calculated the statistical significance of cluster splitting using the R packages ConsensusClusterPlus and sigclust, respectively. Individual cell type composition of these microenvironments was estimated using the DCQ algorithm and a gene expression signature matrix based on the published LM22 leukocyte matrix (Newman et al., 2015) that we augmented with phenotypic profiles of 5 bone marrow-specific cell types (myelomatous plasma cells, plasma memory cells, osteoclasts, osteoblasts, and adipocytes).
Results
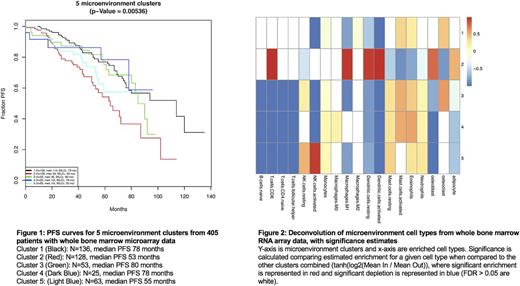
This deconvolution approach accurately estimated the percent of tumor cells in WBM core biopsy as estimated by both microscopy and flow cytometry (PCC = 0.57, RMSE = 10.9%). Unsupervised consensus clustering of whole bone marrow microenvironment gene expression data revealed 5 significantly different subtypes. The two largest subtypes, Cluster 1 (N=136) and Cluster 2 (N=128), had the most divergent outcomes with median PFS values of 78 months and 53 months, respectively. Cluster 1 was significantly enriched for ISS-I patients (FDR < 0.02) while Cluster 2 was enriched for ISS-III patients (FDR < 7x10^-5). Cluster 2 showed a significant increase in estimated M1 macrophages, CD8+ T cells, adipocytes, and dendritic cells with a decrease in estimated osteoclasts versus all other clusters combined (FDRs < 0.05). Cluster 5 (N=63) had the second worst PFS at 55 months, however, it had 44% fewer than expected ISS-III patients (FDR = 0.04). Cluster 5 had fewer estimated T cells and adipocytes, and it was the only cluster with a significant (3-fold) increase in estimated NK cells compared to the other clusters combined (FDRs < 0.05). No clusters were significantly enriched for older individuals, hyperdiploidy or common chromosomal translocations (t(4;14), t(11;14), t(14;16)). No clusters were significantly enriched for CD138+ gene expression-defined high risk patients (UAMS GEP-70) or CD138+ gene expression-based molecular subtypes (Zhan et al., 2006).
Conclusions
We demonstrated the technical feasibility to accurately delineate CD138+ expression profiles from a bulk transcriptome admixture and to use the residual microenvironment transcriptome to cluster patients into distinct microenvironment subtypes. Within these subtypes, the clinical outcomes are not explained by CD138+ molecular features nor ISS stage. For example, Cluster 5 has patients with poor prognosis compared to the rest of the cohort, yet it is not significantly enriched for high risk translocations, ISS-III patients, nor GEP-70 high risk patients. Cluster 5 patients show significant decrease in both CD8+ T cell infiltrate and activated dendritic cells, perhaps reflective of a high risk immune state that would not be responsive to checkpoint inhibition. Moreover, Cluster 5 patients are the only ones to have an increase in NK cells, suggesting one could try to stimulate these NK cells to reinvigorate their anti-tumor immunity. Cluster 2, with the poorest prognosis patients and highest tumor burden (by ISS stage), shows increased CD8+ T cells and dendritic cells (where all other clusters are neutral or decreased in these cell types). Cluster 2 patients may thus benefit from checkpoint blockade, and we plan to further explore relevant checkpoint-related pathways in this cohort. Adding this layer of immune-microenvironment to the analysis of myeloma patient samples enables us to formulate biological hypotheses independent of CD138+ biology and may eventually guide more tailored therapeutic interventions and improve outcomes for patients.
Danziger: Celgene Corporation: Employment. Dervan: Twinstrand Biosciences: Equity Ownership; Celgene Corporation: Employment, Equity Ownership. Schmitz: Celgene Corporation: Employment. Copeland: Celgene Corporation: Employment. Fitch: Celgene Corporation: Employment. Newhall: Celgene Corporation: Employment, Equity Ownership. Barlogie: Celgene Corporation: Consultancy, Research Funding; Millenium Pharmaceuticals: Consultancy, Research Funding. Foy: Celgene Corporation: Employment, Equity Ownership. Trotter: Celgene Corporation: Equity Ownership; Celgene Institute for Translational Research Europe: Employment. Hershberg: Adaptive Biotechnologies: Membership on an entity's Board of Directors or advisory committees; Fraizer Healthcare Partners: Consultancy; NanoString Technologies: Membership on an entity's Board of Directors or advisory committees; Celgene Corporation: Employment, Equity Ownership. Ratushny: Celgene Corporation: Employment. Morgan: Takeda: Consultancy, Honoraria; Bristol Myers: Consultancy, Honoraria; Celgene: Consultancy, Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal